Difference between revisions of "WeightCalculationOfCommonLineSearchByAllSinogram"
(→-M details) |
(→-i1, -12, -i3, -i4 format) |
||
| Line 123: | Line 123: | ||
===-i1, -12, -i3, -i4 format=== | ===-i1, -12, -i3, -i4 format=== | ||
<pre> | <pre> | ||
| − | + | CorFile mrcfile1 mrcfile2 Rot1 Rot2 Mode | |
. . . . . . | . . . . . . | ||
. . . . . . | . . . . . . | ||
</pre> | </pre> | ||
| − | <div> | + | <div>CorFile is used by being put at same directory as File List.</div> |
| − | <div> | + | <div>cf. [[mrcImagesSinogramCorrelation]] about file structure.</div> |
<br> | <br> | ||
Revision as of 07:24, 18 August 2014
WeightCalculationOfCommonLineSearchByAllSinogram is Eos's Command.
Contents
List of option
Main option
| Option | Essential/Optional | Description | Default |
|---|---|---|---|
| -In1 | Optional | Input1: ASCII(File List)(normal correlation) | NULL |
| -In2 | Optional | Input2: ASCII(File List)(derivation1D correlation) | NULL |
| -In3 | Optional | Input3: ASCII(File List)(derivation2D correlation) | NULL |
| -In4 | Optional | Input4: ASCII(File List)(length correlation) | NULL |
| -o | Essential | Output: ASCII | NULL |
| -c | Optional | ConfigurationFile | NULL |
| -m | Optional | Mode | 0 |
| -M | Optional | Mode : choice of used Correlation | 15 |
| -v | Optional | variance of Gaussian weight function | 5 |
| -h | Optional | Help |
-m details
| Value | Description |
|---|---|
| 0 | get memory of mrcImage |
| 1 | don't get memory of mrcImage |
-M details
| Value | Description |
|---|---|
| +1 | ued to -In1 |
| +2 | ued to -In2 |
| +4 | ued to -In3 |
| +8 | ued to -In4 |
| 15 | ued to All Correlation |
-i1, -12, -i3, -i4 format
CorFile mrcfile1 mrcfile2 Rot1 Rot2 Mode
. . . . . .
. . . . . .
Execution example
Input file data
-i1 data
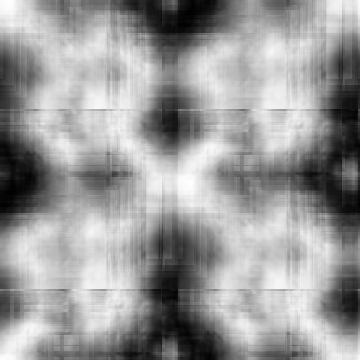
Input-1VOM-2D-sin_Input-1VOM-Rot-2D-sin.Normal.cor Input-1VOM-2D-sin.mrc Input-1VOM-Rot-2D-sin.mrc 1.570796 1.570796 1 Input-1VOM-2D-sin_Input-1VOM-Rot1-2D-sin.Normal.cor Input-1VOM-2D-sin.mrc Input-1VOM-Rot1-2D-sin.mrc 0.000000 0.000000 z-rotation Input-1VOM-Rot-2D-sin_Input-1VOM-Rot1-2D-sin.Normal.cor Input-1VOM-Rot-2D-sin.mrc Input-1VOM-Rot1-2D-sin.mrc 1.570796 1.570796 1
 |
Min Max |
-0.925733 (174, 223, 0) 0.986152 (271, 281, 0) |
 |
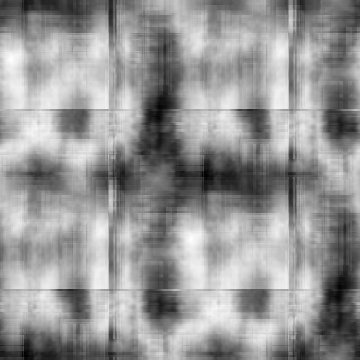
Min Max |
-0.969656 (10, 168, 0) 0.96977 (10, 357, 0) |
 |
Min Max |
-0.79716 (206, 183, 0) 0.986575 (190, 120, 0) |
-i2 data
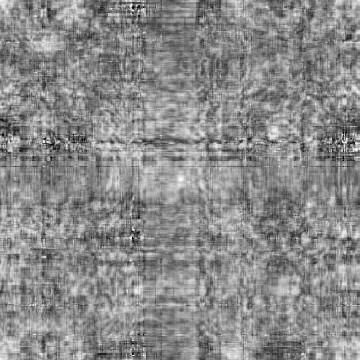
Input-1VOM-2D-sin_Input-1VOM-Rot-2D-sin.Derivation1D.cor Input-1VOM-2D-sin.mrc Input-1VOM-Rot-2D-sin.mrc 1.570796 1.570796 1 Input-1VOM-2D-sin_Input-1VOM-Rot1-2D-sin.Derivation1D.cor Input-1VOM-2D-sin.mrc Input-1VOM-Rot1-2D-sin.mrc 0.000000 0.000000 z-rotation Input-1VOM-Rot-2D-sin_Input-1VOM-Rot1-2D-sin.Derivation1D.cor Input-1VOM-Rot-2D-sin.mrc Input-1VOM-Rot1-2D-sin.mrc 1.570796 1.570796 1
 |
Min Max |
-0.999642 (131, 329, 0) 0.921779 (132, 73, 0) |
 |
Min Max |
-0.995358 (132, 121, 0) 0.991247 (133, 4, 0) |
 |
Min Max |
-0.98175 (356, 217, 0) 0.97865 (0, 217, 0) |
-i3 data
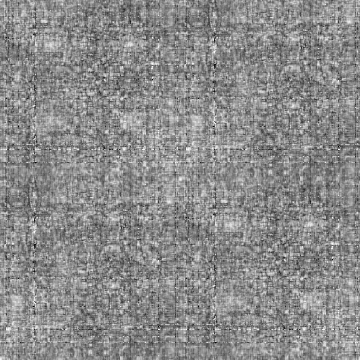
Input-1VOM-2D-sin_Input-1VOM-Rot-2D-sin.Derivation2D.cor Input-1VOM-2D-sin.mrc Input-1VOM-Rot-2D-sin.mrc 1.570796 1.570796 1 Input-1VOM-2D-sin_Input-1VOM-Rot1-2D-sin.Derivation2D.cor Input-1VOM-2D-sin.mrc Input-1VOM-Rot1-2D-sin.mrc 0.000000 0.000000 z-rotation Input-1VOM-Rot-2D-sin_Input-1VOM-Rot1-2D-sin.Derivation2D.cor Input-1VOM-Rot-2D-sin.mrc Input-1VOM-Rot1-2D-sin.mrc 1.570796 1.570796 1
 |
Min Max |
-1 (285, 75, 0) 1 (285, 61, 0) |
 |
Min Max |
-1 (158, 33, 0) 1 (158, 30, 0) |
 |
Min Max |
-0.970911 (132, 30, 0) 1 (135, 30, 0) |
-i4 data
Input-1VOM-2D-sin_Input-1VOM-Rot-2D-sin.Length.cor Input-1VOM-2D-sin.mrc Input-1VOM-Rot-2D-sin.mrc 1.570796 1.570796 1 Input-1VOM-2D-sin_Input-1VOM-Rot1-2D-sin.Length.cor Input-1VOM-2D-sin.mrc Input-1VOM-Rot1-2D-sin.mrc 0.000000 0.000000 z-rotation Input-1VOM-Rot-2D-sin_Input-1VOM-Rot1-2D-sin.Length.cor Input-1VOM-Rot-2D-sin.mrc Input-1VOM-Rot1-2D-sin.mrc 1.570796 1.570796 1
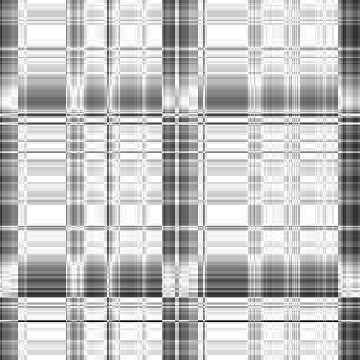 |
Min Max |
0.778626 (27, 38, 0) 1 (27, 0, 0) |
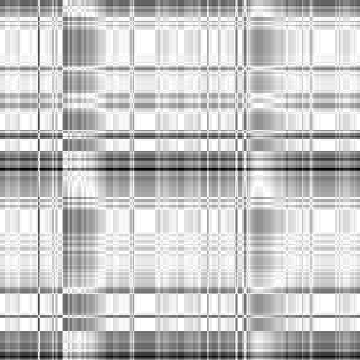 |
Min Max |
0.699187 (27, 191, 0) 1 (0, 0, 0) |
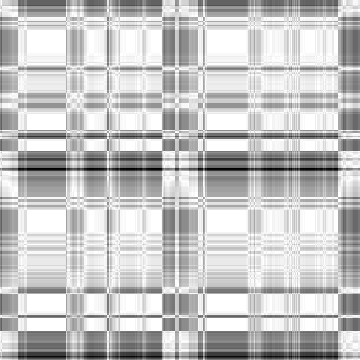 |
Min Max |
0.699187 (0, 191, 0) 1 (67, 0, 0) |
Case: Options only essential
Input data get end : CommonLinePositionData Input data get end : CommonLinePositionData Input data get end : CommonLinePositionData Input data get end : CommonLinePositionData weight calculation start deltaweight = 1.414214 FinalCorrelationMin = 10.000000 weight[0][1.414214, 8.000000] >>>> weightMinOfCorrelation = 0.000000 weight[1][1.414214, 8.000000] >>>> weightMinOfCorrelation = 0.000000 weight[2][1.414214, 8.000000] >>>> weightMinOfCorrelation = 0.000000 weight[3][1.414214, 8.000000] >>>> weightMinOfCorrelation = 0.000000 deltaweight = 1.189207 FinalCorrelationMin = 3.000000 weight[0][0.000000, 0.000000] >>>> weightMinOfCorrelation = 0.000000 weight[1][0.000000, 0.000000] >>>> weightMinOfCorrelation = 0.000000 weight[2][0.000000, 0.000000] >>>> weightMinOfCorrelation = 0.000000 weight[3][0.000000, 0.000000] >>>> weightMinOfCorrelation = 0.000000 deltaweight = 1.090508 FinalCorrelationMin = 3.000000 weight[0][0.000000, 0.000000] >>>> weightMinOfCorrelation = 0.000000 weight[1][0.000000, 0.000000] >>>> weightMinOfCorrelation = 0.000000 weight[2][0.000000, 0.000000] >>>> weightMinOfCorrelation = 0.000000 weight[3][0.000000, 0.000000] >>>> weightMinOfCorrelation = 0.000000 ... deltaweight = 1.000000 FinalCorrelationMin = 3.000000 weight[0][0.000000, 0.000000] >>>> weightMinOfCorrelation = 0.000000 weight[1][0.000000, 0.000000] >>>> weightMinOfCorrelation = 0.000000 weight[2][0.000000, 0.000000] >>>> weightMinOfCorrelation = 0.000000 weight[3][0.000000, 0.000000] >>>> weightMinOfCorrelation = 0.000000 deltaweight = 1.000000 FinalCorrelationMin = 3.000000 weight[0][0.000000, 0.000000] >>>> weightMinOfCorrelation = 0.000000 weight[1][0.000000, 0.000000] >>>> weightMinOfCorrelation = 0.000000 weight[2][0.000000, 0.000000] >>>> weightMinOfCorrelation = 0.000000 weight[3][0.000000, 0.000000] >>>> weightMinOfCorrelation = 0.000000
Weight of I1 is 0.000000 Weight of I2 is 0.000000 Weight of I3 is 0.000000 Weight of I4 is 0.000000
Option -M
Case: M=3
Case: No other settings
Weight of I1 is 0.771105 Weight of I2 is 3.459215
Case: v=1
Weight of I1 is 0.840896 Weight of I2 is 3.772301
Case: m=1
Input data get end : CommonLinePositionData Input data get end : CommonLinePositionData weight calculation start deltaweight = 1.414214 FinalCorrelationMin = 2.628996 weight[0][1.414214, 8.000000] >>>> weightMinOfCorrelation = 1.414214 weight[1][1.414214, 8.000000] >>>> weightMinOfCorrelation = 7.071068 deltaweight = 1.189207 FinalCorrelationMin = 2.616069 weight[0][1.000000, 2.000000] >>>> weightMinOfCorrelation = 1.000000 weight[1][5.000000, 10.000000] >>>> weightMinOfCorrelation = 9.756828 deltaweight = 1.090508 FinalCorrelationMin = 2.612862 weight[0][0.840896, 1.189207] >>>> weightMinOfCorrelation = 0.840896 weight[1][8.204482, 11.602890] >>>> weightMinOfCorrelation = 11.476005 ... deltaweight = 1.000000 FinalCorrelationMin = 2.612196 weight[0][0.771105, 0.771105] >>>> weightMinOfCorrelation = 0.771105 weight[1][11.567817, 11.567817] >>>> weightMinOfCorrelation = 11.567817 deltaweight = 1.000000 FinalCorrelationMin = 2.612196 weight[0][0.771105, 0.771105] >>>> weightMinOfCorrelation = 0.771105 weight[1][11.567817, 11.567817] >>>> weightMinOfCorrelation = 11.567817 deltaweight = 1.000000 FinalCorrelationMin = 2.612196 weight[0][0.771105, 0.771105] >>>> weightMinOfCorrelation = 0.771105 weight[1][11.567817, 11.567817] >>>> weightMinOfCorrelation = 11.567817
Weight of I1 is 0.771105 Weight of I2 is 11.567817
Case: M=5
Weight of I1 is 0.000000 Weight of I2 is 0.000000
Case: M=6
Weight of I1 is 0.000000 Weight of I2 is 0.000000
Case: M=7
Weight of I1 is 0.000000 Weight of I2 is 0.000000 Weight of I3 is 0.000000
Case: M=9
Weight of I1 is 0.840327 Weight of I2 is 3.769748
Case: M=10
Weight of I1 is 0.840327 Weight of I2 is 3.769748
Case: M=11
Weight of I1 is 0.766941 Weight of I2 is 0.766941 Weight of I3 is 11.505344
Case: M=12
Weight of I1 is 0.000000 Weight of I2 is 0.000000
Case: M=13
Weight of I1 is 0.000000 Weight of I2 is 0.000000 Weight of I3 is 0.000000
Case: M=14
Weight of I1 is 0.766941 Weight of I2 is 0.766941 Weight of I3 is 11.505344