Difference between revisions of "pdbAtomSection"
From EosPedia
(Created page with "'''pdbAtomSection''' is Eos's Command that cuts out section from Atoms Model, and outputs to ps file. == List of option == === Main option === <table border="1">...") |
(→Main option) |
||
| Line 38: | Line 38: | ||
<td>-zmin</td> | <td>-zmin</td> | ||
<td>Optional</td> | <td>Optional</td> | ||
| − | <td> | + | <td>z min</td> |
<td>0.0</td> | <td>0.0</td> | ||
</tr> | </tr> | ||
| Line 44: | Line 44: | ||
<td>-zmax</td> | <td>-zmax</td> | ||
<td>Optional</td> | <td>Optional</td> | ||
| − | <td> | + | <td>z max</td> |
<td>90.0</td> | <td>90.0</td> | ||
</tr> | </tr> | ||
| Line 56: | Line 56: | ||
<td>-r</td> | <td>-r</td> | ||
<td>Optional</td> | <td>Optional</td> | ||
| − | <td> | + | <td>radius</td> |
<td>1.0</td> | <td>1.0</td> | ||
</tr> | </tr> | ||
| Line 62: | Line 62: | ||
<td>-clw</td> | <td>-clw</td> | ||
<td>Optional</td> | <td>Optional</td> | ||
| − | <td> | + | <td>Width of Circle</td> |
<td>0.1</td> | <td>0.1</td> | ||
</tr> | </tr> | ||
| Line 68: | Line 68: | ||
<td>-plw</td> | <td>-plw</td> | ||
<td>Optional</td> | <td>Optional</td> | ||
| − | <td> | + | <td>Width of Peptide bond</td> |
<td>0.1</td> | <td>0.1</td> | ||
</tr> | </tr> | ||
| Line 119: | Line 119: | ||
<td> </td> | <td> </td> | ||
</tr> | </tr> | ||
| − | </table> | + | </table> |
=== -m details === | === -m details === | ||
Revision as of 06:10, 7 August 2014
pdbAtomSection is Eos's Command that cuts out section from Atoms Model, and outputs to ps file.
Contents
- 1 List of option
- 2 Execution example
List of option
Main option
| Option | Essential/Optional | Description | Default |
|---|---|---|---|
| -i | Essential | Input: PDB | NULL |
| -if | Optional | Input: FlagsFile | NULL |
| -o | Essential | Output: psfile | NULL |
| -dist | Optional | Distance of Sections | 2.5 |
| -zmin | Optional | z min | 0.0 |
| -zmax | Optional | z max | 90.0 |
| -fm | Optional | flag mode | 13 |
| -r | Optional | radius | 1.0 |
| -clw | Optional | Width of Circle | 0.1 |
| -plw | Optional | Width of Peptide bond | 0.1 |
| -fontsize | Optional | font size | 2.0 |
| -AS | Optional | absolute scale of PS file (-AS 1 := 1 mm/A) | 1.0 |
| -shiftx | Optional | pdb Shift x [A] | 0.0 |
| -shifty | Optional | pdb Shift y [A] | 0.0 |
| -shiftz | Optional | pdb Shift z [A] | 0.0 |
| -c | Optional | ConfigurationFile | NULL |
| -m | Optional | Mode | 0 |
| -h | Optional | Help |
-m details
| Value | Description |
|---|---|
| 0 |
-fm details
| Value | Description |
|---|---|
| 0 | display nothing |
| 1 | one char amino acid name |
| 2 | three char amino acid name |
| 4 | chain identifier |
| 8 | sequence number |
| 16 | display any atom |
-if format
chainIdertifier&sequenceNumber(R123) FlagMode circleRedius circlelinewidth fontsize
Execution example
Input file image
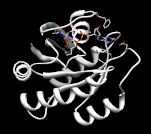 |
Gravity Centre(CA) Atom far from Centre(CA) |
6.053012e+00 2.519590e+01 1.189075e+01 3.286664e+01 |
Example of options only essential
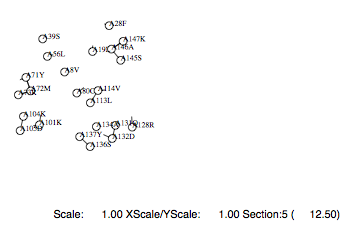 |
|---|
| 6/90 Page (Left bottom part) |
Option -AS
Case: AS=5
 |
|---|
| 6/90 Page |
In the following, Setting at AS=5
Option -shiftx
Case: shiftx=10
 |
|---|
| 6/90 Page |
In the following, Setting at shiftx=10
Option -shifty
Case: shifty=10
 |
|---|
| 6/90 Page |
Option -shiftz
Case: shiftz=1
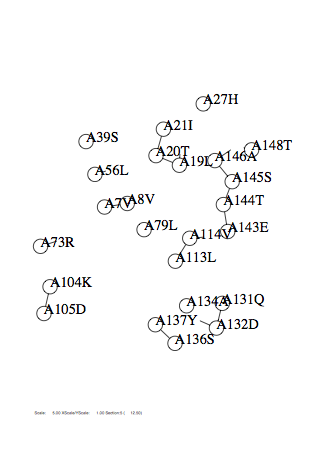 |
|---|
| 6/90 Page |
Option -dist
Case: dist=3
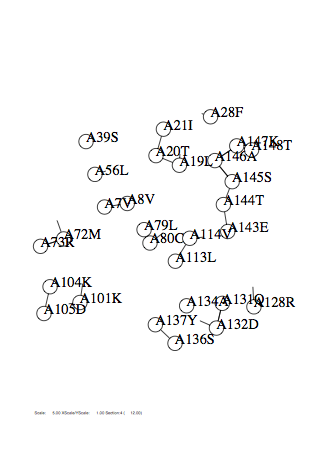 |
|---|
| 5/90 Page |
Option -zmin
Case: zmin=1
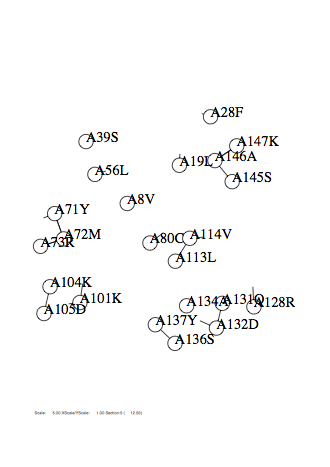 |
|---|
| 5/89 Page |
Option -zmax
Case: zmax=15
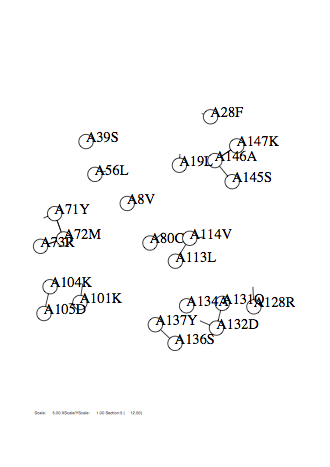 |
|---|
| 6/15 Page |
Option -r
Case: r=0.5
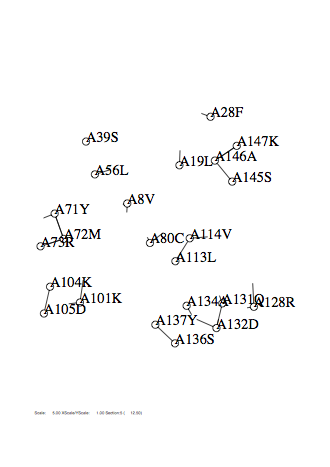 |
|---|
| 6/90 Page |
Option -plw
Case: plw=0.3
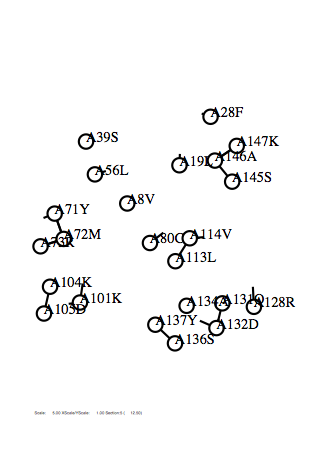 |
|---|
| 6/90 Page |
Option -fontsize
Case: fontsize=1
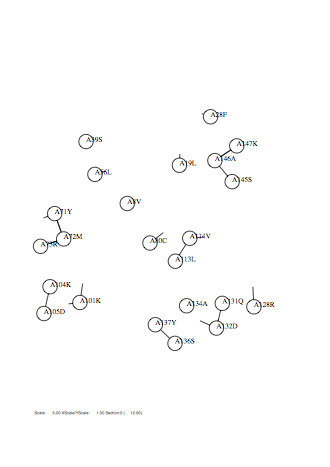 |
|---|
| 6/90 Page |
Option -fm
Case: fm=0
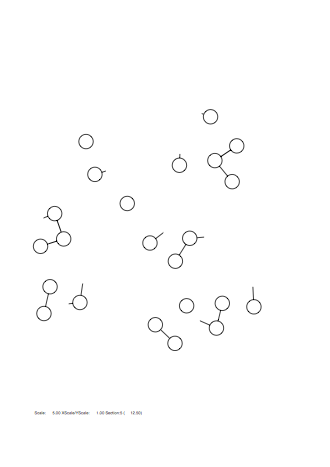 |
|---|
| 6/90 Page |
Case: fm=1
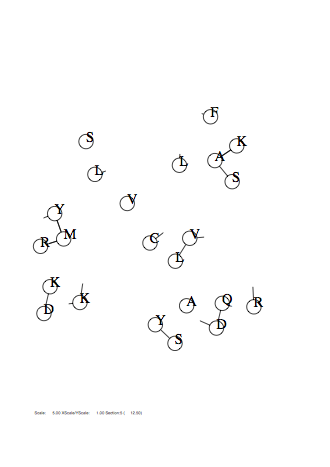 |
|---|
| 6/90 Page |
Case: fm=2
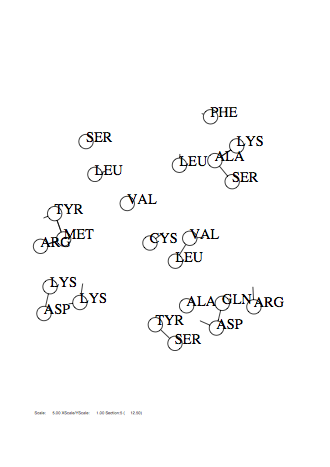 |
|---|
| 6/90 Page |
Case: fm=4
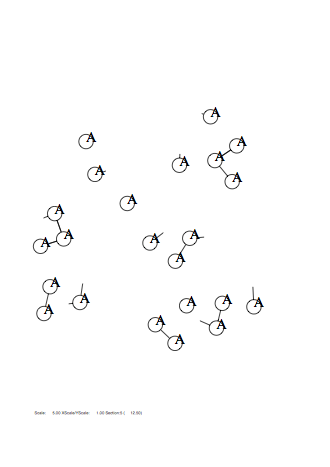 |
|---|
| 6/90 Page |
Case: fm=8
 |
|---|
| 6/90 Page |
Case: fm=16
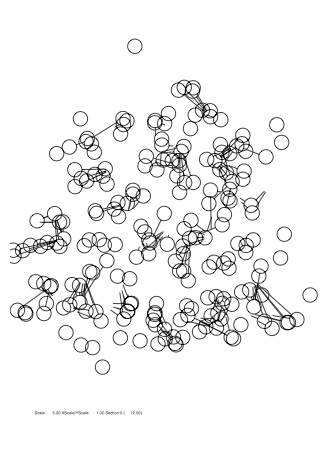 |
|---|
| 6/90 Page |
Option -if
-if 's data
A28 0 2.0 0.3 2 A147 1 0.5 0.3 4 A114 2 1.0 0.3 1 A128 4 1.0 0.3 4 A8 8 1.0 0.3 4 A56 16 0.5 0.3 4 A39 31 0.7 0.3 3
Output
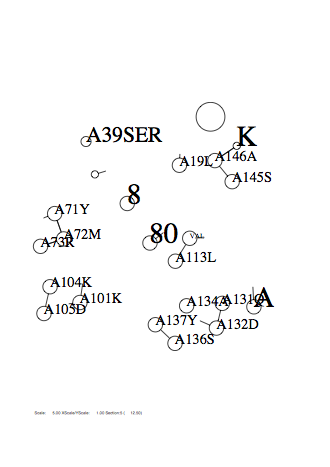 |
|---|
| 6/90 Page |