mrcImagesSinogramCorrelation
mrcImagesSinogramCorrelation is Eos's Command that performs correlation among multiple Sinograms. Output file is used by WeightCalculationOfCommonLineSearchByAllSinogram.
Contents
List of option
Main option
| Option | Essential/Optional | Description | Default |
|---|---|---|---|
| -I | Essential | Input: ASCII(FileList) | NULL |
| -oH | Optional | OutputDataFileHeader | test |
| -Rd | Optional | output results directory name | Result |
| -Nd | Optional | irectory name of output normalcorrelation in results directory(Create in -Rd) | Normal |
| -D1d | Optional | directory name of output derivation1Dcorrelation in results directory(Create in -Rd) | Derivation1D |
| -D2d | Optional | directory name of output derivation2Dcorrelation in results directory(Create in -Rd) | Derivation2D |
| -Ld | Optional | directory name of output lengthcorrelation in results directory(Create in -Rd) | Length |
| -CE | Optional | output correlation file extension | cor |
| -LE | Optional | output file list extension | list |
| -Nm | Optional | NormalCorrelationMode | 0 |
| -D1m | Optional | Derivation1DCorrelationMode | 0 |
| -D2m | Optional | Derivation2DCorrelationMode | 0 |
| -Lm | Optional | LengthCorrelationMode | 0 |
| -LTM | Optional | LengthThresholdMode | 0 |
| -LTR | Optional | LengthThresholdRatio | 0.25 |
| -CM | Optional | CreateFileMode | 0 |
| -c | Optional | ConfigurationFile | NULL |
| -m | Optional | Mode | 0 |
| -h | Optional | Help |
-m details
| Value | Description |
|---|---|
| 0 |
-Nm details
| Value | Description |
|---|---|
| 0 | create normal correlation(-Nd (Specify Output Directory) |
| 1 | not create normal correlation |
-D1m details
| Value | Description |
|---|---|
| 0 | create derivation1D correlation(-D1d (Specify Output Directory) |
| 1 | not create derivation1D correlation |
-D2m details
| Value | Description |
|---|---|
| 0 | create derivation2D correlation(-D2d (Specify Output Directory) |
| 1 | not create derivation2D correlation |
-Lm details
| Value | Description |
|---|---|
| 0 | create length correlation (-Ld(Specify Output Directory), -LTM, -LTR) |
| 1 | not creat length correlation |
-CM details
| Value | Description |
|---|---|
| 0 | Output image of correlation and list |
| 1 | Output list only |
-I format
file1, EA1 Rot1-1 Rot1-2 Rot1-3
file2, EA2 Rot2-1 Rot2-2 Rot2-3
file3, EA3 Rot3-1 Rot3-2 Rot3-3
. , . . . .
. , . . . .
. , . . . .
file名の先頭を/にすると命名規則により相関fileを作成できないので注意。
Created Directory and File
Directory
Rd Rd/Nd (D1d, D2d, Ld) Example: Result/Normal/
ListFile
Rd/Nd/oH-Nd.LE Example: Result/Normal/test-Normal.list
CorrelationFile
Rd/Nd/infile1_infile2.Nd.CE Example: Result/Normal/test1_test2.Normal.cor
Execution example
Input file's data
Input-1VOM-2D-sin.mrc, YOYS 0 0 0 Input-1VOM-Rot-2D-sin.mrc, YOYS 90 0 0 Input-1VOM-Rot3-2D-sin.mrc, YOYS 0 90 0
 |
Min Max |
-3045.85 (79, 268, 0) 1.53325e+06 (27, 1, 0) |
 |
Min Max |
-16429.6 (78, 357, 0) 1.31372e+06 (44, 38, 0) |
 |
Min Max |
-1732.14 (3, 3, 0) 1.39371e+06 (51, 202, 0) |
Case: Options only essential
LengthCorrelation=0.840580 InLength[0]=58 RefLength[0]=80 LengthCorrelation=0.840580 InLength[0]=58 RefLength[1]=80 LengthCorrelation=0.840580 InLength[0]=58 RefLength[2]=80 LengthCorrelation=0.840580 InLength[0]=58 RefLength[3]=80 LengthCorrelation=0.840580 InLength[0]=58 RefLength[4]=80 LengthCorrelation=0.872180 InLength[0]=58 RefLength[5]=75 LengthCorrelation=0.872180 InLength[0]=58 RefLength[6]=75 ... LengthCorrelation=0.852941 InLength[359]=58 RefLength[355]=78 LengthCorrelation=0.852941 InLength[359]=58 RefLength[356]=78 LengthCorrelation=0.840580 InLength[359]=58 RefLength[357]=80 LengthCorrelation=0.840580 InLength[359]=58 RefLength[358]=80 LengthCorrelation=0.840580 InLength[359]=58 RefLength[359]=80 LengthCorrelation=1.000000 InLength[0]=58 RefLength[0]=58 LengthCorrelation=0.945455 InLength[0]=58 RefLength[1]=52 LengthCorrelation=0.905660 InLength[0]=58 RefLength[2]=48 LengthCorrelation=0.862745 InLength[0]=58 RefLength[3]=44 LengthCorrelation=0.925926 InLength[0]=58 RefLength[4]=50 ... LengthCorrelation=0.973451 InLength[359]=58 RefLength[355]=55 LengthCorrelation=0.973451 InLength[359]=58 RefLength[356]=55 LengthCorrelation=0.973451 InLength[359]=58 RefLength[357]=55 LengthCorrelation=0.973451 InLength[359]=58 RefLength[358]=55 LengthCorrelation=0.973451 InLength[359]=58 RefLength[359]=55 LengthCorrelation=0.840580 InLength[0]=80 RefLength[0]=58 LengthCorrelation=0.787879 InLength[0]=80 RefLength[1]=52 LengthCorrelation=0.750000 InLength[0]=80 RefLength[2]=48 LengthCorrelation=0.709677 InLength[0]=80 RefLength[3]=44 LengthCorrelation=0.769231 InLength[0]=80 RefLength[4]=50 ... LengthCorrelation=0.840580 InLength[359]=80 RefLength[350]=58 LengthCorrelation=0.881119 InLength[359]=80 RefLength[351]=63 LengthCorrelation=0.881119 InLength[359]=80 RefLength[352]=63 LengthCorrelation=0.873239 InLength[359]=80 RefLength[353]=62 LengthCorrelation=0.796992 InLength[359]=80 RefLength[354]=53 LengthCorrelation=0.814815 InLength[359]=80 RefLength[355]=55 LengthCorrelation=0.814815 InLength[359]=80 RefLength[356]=55 LengthCorrelation=0.814815 InLength[359]=80 RefLength[357]=55 LengthCorrelation=0.814815 InLength[359]=80 RefLength[358]=55 LengthCorrelation=0.814815 InLength[359]=80 RefLength[359]=55
In the case of default setting, file structure is as the following.
$EOS_HOME/Result/Derivation1D/Input-1VOM-2D-sin_Input-1VOM-Rot-2D-sin.Derivation1D.cor $EOS_HOME/Result/Derivation1D/Input-1VOM-2D-sin_Input-1VOM-Rot3-2D-sin.Derivation1D.cor $EOS_HOME/Result/Derivation1D/Input-1VOM-Rot-2D-sin_Input-1VOM-Rot3-2D-sin.Derivation1D.cor $EOS_HOME/Result/Derivation1D/test-Derivation1D.list $EOS_HOME/Result/Derivation2D/Input-1VOM-2D-sin_Input-1VOM-Rot-2D-sin.Derivation2D.cor $EOS_HOME/Result/Derivation2D/Input-1VOM-2D-sin_Input-1VOM-Rot3-2D-sin.Derivation2D.cor $EOS_HOME/Result/Derivation2D/Input-1VOM-Rot-2D-sin_Input-1VOM-Rot3-2D-sin.Derivation2D.cor $EOS_HOME/Result/Derivation2D/test-Derivation2D.list $EOS_HOME/Result/Length/Input-1VOM-2D-sin_Input-1VOM-Rot-2D-sin.Length.cor $EOS_HOME/Result/Length/Input-1VOM-2D-sin_Input-1VOM-Rot3-2D-sin.Length.cor $EOS_HOME/Result/Length/Input-1VOM-Rot-2D-sin_Input-1VOM-Rot3-2D-sin.Length.cor $EOS_HOME/Result/Length/test-Length.list $EOS_HOME/Result/Normal/Input-1VOM-2D-sin_Input-1VOM-Rot-2D-sin.Normal.cor $EOS_HOME/Result/Normal/Input-1VOM-2D-sin_Input-1VOM-Rot3-2D-sin.Normal.cor $EOS_HOME/Result/Normal/Input-1VOM-Rot-2D-sin_Input-1VOM-Rot3-2D-sin.Normal.cor $EOS_HOME/Result/Normal/test-Normal.list
Option -Nm
Case: Nm=0(Default)
 |
Min Max |
-0.925733 (174, 223, 0) 0.986152 (271, 281, 0) |
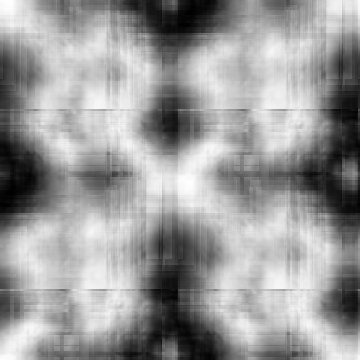 |
Min Max |
-0.969656 (10, 168, 0) 0.96977 (10, 357, 0) |
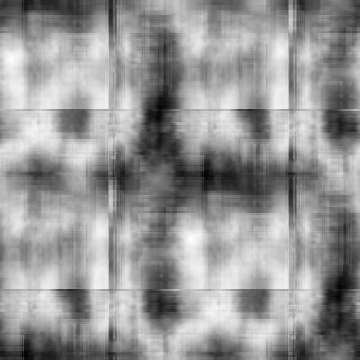 |
Min Max |
-0.79716 (206, 183, 0) 0.986575 (190, 120, 0) |
Input-1VOM-2D-sin_Input-1VOM-Rot-2D-sin.Normal.cor Input-1VOM-2D-sin.mrc Input-1VOM-Rot-2D-sin.mrc 1.570796 1.570796 1 Input-1VOM-2D-sin_Input-1VOM-Rot3-2D-sin.Normal.cor Input-1VOM-2D-sin.mrc Input-1VOM-Rot3-2D-sin.mrc 0.000000 0.000000 1 Input-1VOM-Rot-2D-sin_Input-1VOM-Rot3-2D-sin.Normal.cor Input-1VOM-Rot-2D-sin.mrc Input-1VOM-Rot3-2D-sin.mrc 0.000000 4.712389 1
Option -D1d
Case: D1d=0(Default)
 |
Min Max |
-0.999642 (131, 329, 0) 0.921779 (132, 73, 0) |
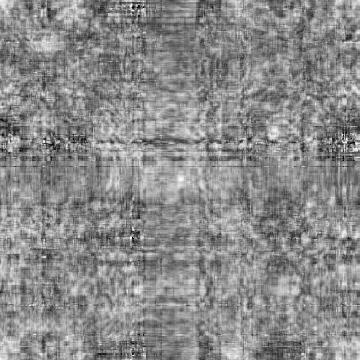 |
Min Max |
-0.995358 (132, 121, 0) 0.991247 (133, 4, 0) |
 |
Min Max |
-0.98175 (356, 217, 0) 0.97865 (0, 217, 0) |
Input-1VOM-2D-sin_Input-1VOM-Rot-2D-sin.Derivation1D.cor Input-1VOM-2D-sin.mrc Input-1VOM-Rot-2D-sin.mrc 1.570796 1.570796 1 Input-1VOM-2D-sin_Input-1VOM-Rot3-2D-sin.Derivation1D.cor Input-1VOM-2D-sin.mrc Input-1VOM-Rot3-2D-sin.mrc 0.000000 0.000000 1 Input-1VOM-Rot-2D-sin_Input-1VOM-Rot3-2D-sin.Derivation1D.cor Input-1VOM-Rot-2D-sin.mrc Input-1VOM-Rot3-2D-sin.mrc 0.000000 4.712389 1
Option -D2d
Case: D2d=0(Default)
 |
Min Max |
-1 (285, 75, 0) 1 (285, 61, 0) |
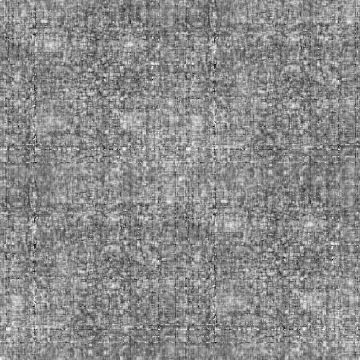 |
Min Max |
-1 (158, 33, 0) 1 (158, 30, 0) |
 |
Min Max |
-0.970911 (132, 30, 0) 1 (135, 30, 0) |
Input-1VOM-2D-sin_Input-1VOM-Rot-2D-sin.Derivation2D.cor Input-1VOM-2D-sin.mrc Input-1VOM-Rot-2D-sin.mrc 1.570796 1.570796 1 Input-1VOM-2D-sin_Input-1VOM-Rot3-2D-sin.Derivation2D.cor Input-1VOM-2D-sin.mrc Input-1VOM-Rot3-2D-sin.mrc 0.000000 0.000000 1 Input-1VOM-Rot-2D-sin_Input-1VOM-Rot3-2D-sin.Derivation2D.cor Input-1VOM-Rot-2D-sin.mrc Input-1VOM-Rot3-2D-sin.mrc 0.000000 4.712389 1
Option -Ld
Case: Ld=0(Default)
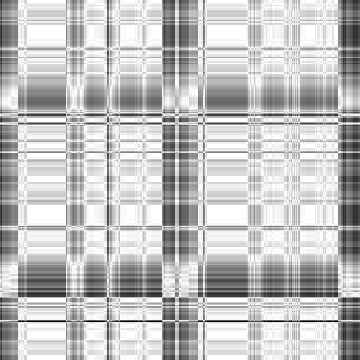 |
Min Max |
0.778626 (27, 38, 0) 1 (27, 0, 0) |
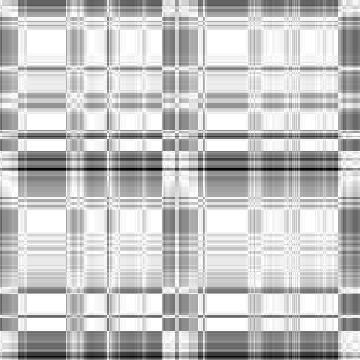 |
Min Max |
0.699187 (27, 191, 0) 1 (0, 0, 0) |
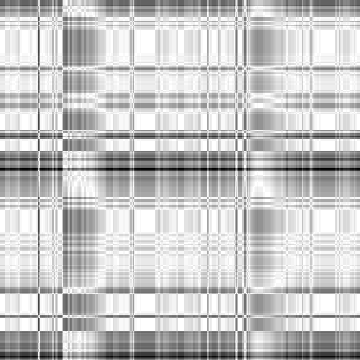 |
Min Max |
0.699187 (0, 191, 0) 1 (67, 0, 0) |
Input-1VOM-2D-sin_Input-1VOM-Rot-2D-sin.Length.cor Input-1VOM-2D-sin.mrc Input-1VOM-Rot-2D-sin.mrc 1.570796 1.570796 1 Input-1VOM-2D-sin_Input-1VOM-Rot3-2D-sin.Length.cor Input-1VOM-2D-sin.mrc Input-1VOM-Rot3-2D-sin.mrc 0.000000 0.000000 1 Input-1VOM-Rot-2D-sin_Input-1VOM-Rot3-2D-sin.Length.cor Input-1VOM-Rot-2D-sin.mrc Input-1VOM-Rot3-2D-sin.mrc 0.000000 4.712389 1
Option -LTR
Case: LTR=0.5
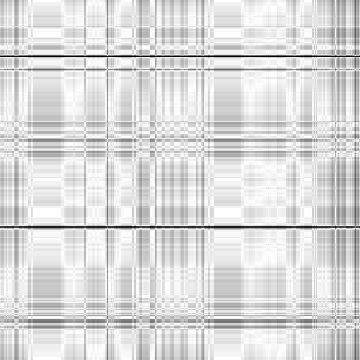 |
Min Max |
0.0487805 (28, 303, 0) 1 (28, 0, 0) |
 |
Min Max |
0.16092 (28, 68, 0) 1 (1, 0, 0) |
 |
Min Max |
0.0487805 (303, 31, 0) 1 (31, 0, 0) |
Input-1VOM-2D-sin_Input-1VOM-Rot-2D-sin.Length.cor Input-1VOM-2D-sin.mrc Input-1VOM-Rot-2D-sin.mrc 1.570796 1.570796 1 Input-1VOM-2D-sin_Input-1VOM-Rot3-2D-sin.Length.cor Input-1VOM-2D-sin.mrc Input-1VOM-Rot3-2D-sin.mrc 0.000000 0.000000 z-rotation Input-1VOM-Rot-2D-sin_Input-1VOM-Rot3-2D-sin.Length.cor Input-1VOM-Rot-2D-sin.mrc Input-1VOM-Rot3-2D-sin.mrc 1.570796 1.570796 1